Module 3: Data Preprocessing and Features Selection#
Welcome to Module 3 of our learning journey! In this segment, we’ll delve into the critical aspects of Data Preprocessing and Feature Selection.
As aspiring data enthusiasts, you’ve likely realized that the journey from raw data to meaningful insights involves careful preparation and strategic selection. This module is designed to equip you with the essential skills to transform raw, messy data into a refined and feature-rich dataset, setting the stage for robust machine learning models.
Key Objectives:#
Data cleaning and imputation are essential steps in the data preprocessing pipeline for several reasons:
Handling Missing Data:
Real-world datasets often contain missing values due to various reasons such as measurement errors, system failures, or human error. Data cleaning and imputation techniques address these gaps, ensuring a complete and usable dataset.
Ensuring Data Quality:
Inaccurate or inconsistent data can significantly impact the performance and reliability of machine learning models. Cleaning the data involves identifying and rectifying errors, ensuring the overall quality and integrity of the dataset.
Mitigating Outliers:
Outliers, or extreme values, can distort statistical analyses and model predictions. Data cleaning involves identifying and handling outliers appropriately, preventing them from unduly influencing the model’s behavior.
Improving Model Performance:
Machine learning algorithms often struggle with missing values and outliers. By cleaning and imputing data, the dataset becomes more suitable for model training, leading to improved performance and generalization on new, unseen data.
Enhancing Data Interpretability:
Clean and well-imputed data fosters a better understanding of the underlying patterns and relationships. Researchers and analysts can trust the results and interpretations derived from a dataset that has undergone thorough cleaning and imputation.
Meeting Assumptions of Statistical Methods:
Many statistical methods assume certain characteristics of the data, such as normal distribution or absence of missing values. Data cleaning ensures that the dataset adheres to these assumptions, allowing for the accurate application of statistical techniques.
Building Trust in Results:
Stakeholders, decision-makers, and users of the machine learning model rely on accurate and trustworthy results. Data cleaning and imputation contribute to the credibility of the model’s outcomes, fostering trust in the decision-making process.
Reducing Bias and Error:
Incomplete or biased datasets can lead to skewed and inaccurate model predictions. Data cleaning minimizes bias, ensuring that the model is trained on a representative and unbiased set of features.
Why Data Preprocessing Matters:#
Raw data seldom fits neatly into the algorithms we love to employ. Data preprocessing is the unsung hero that transforms raw data into a well-behaved companion, ensuring your models can extract meaningful patterns and insights.
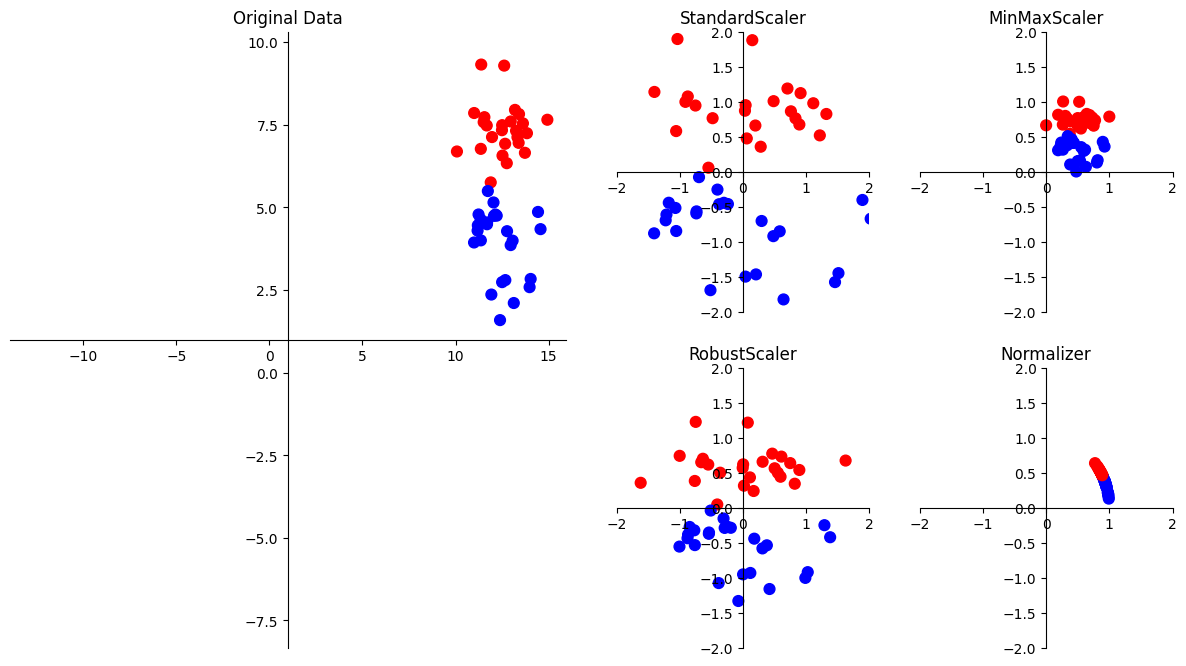
Visualising scaling methods#
Let us visulaise what does scaling methods do to the input data. The following are some of the popular preprocessing methods in the sklearn library.
The four preprocessing methods—StandardScaler, MinMaxScaler, Normalizer, and RobustScaler—from scikit-learn’s preprocessing module serve different purposes in preparing and scaling data for machine learning models. Here’s an overview of each:
StandardScaler:
Method:
StandardScalerstandardizes features by removing the mean and scaling to unit variance.Usage: It transforms features so that they have a mean of 0 and a standard deviation of 1.
When to Use: Suitable for algorithms that assume features follow a Gaussian distribution and for ensuring consistent scales for features.
MinMaxScaler:
Method:
MinMaxScalerscales features to a specified range, typically between 0 and 1.Usage: It transforms features, maintaining the original data’s shape while scaling it to a specified range.
When to Use: Useful when features need to be on a similar scale, but you want to preserve the original distribution.
Normalizer:
Method:
Normalizerscales each sample (row) independently, such that the Euclidean norm (L2 norm) of each row is equal to 1.Usage: It normalizes samples to a unit norm, useful when the direction of each data point is more important than its magnitude.
When to Use: Appropriate when working with datasets where the magnitude of the individual samples is crucial.
RobustScaler:
Method:
RobustScalerscales features using statistics that are robust to outliers. It uses the median and the interquartile range (IQR).Usage: It scales features while being less affected by outliers compared to
StandardScaler.When to Use: Suitable when the dataset contains outliers and a more robust scaling method is needed.
In summary, these preprocessing methods provide flexibility in handling different types of data and scaling requirements. The choice of scaler depends on the characteristics of the data and the assumptions of the machine learning algorithm being used.
In practice, StandardScaler is the most typical approach as it requires less assumptions and works well in most scenarios.
!pip install kaleido
Collecting kaleido
Downloading kaleido-0.2.1-py2.py3-none-manylinux1_x86_64.whl (79.9 MB)
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 79.9/79.9 MB 7.5 MB/s eta 0:00:00m eta 0:00:01[36m0:00:01
?25hInstalling collected packages: kaleido
Successfully installed kaleido-0.2.1
import matplotlib.pyplot as plt
import numpy as np
from sklearn.datasets import make_blobs
from sklearn.preprocessing import StandardScaler, MinMaxScaler, Normalizer, RobustScaler
cm2 = "bwr"
X, y = make_blobs(n_samples=50, centers=2, random_state=4, cluster_std=1)
X += 3
plt.figure(figsize=(15, 8))
main_ax = plt.subplot2grid((2, 4), (0, 0), rowspan=2, colspan=2)
main_ax.scatter(X[:, 0], X[:, 1], c=y, cmap=cm2, s=60)
maxx = np.abs(X[:, 0]).max()
maxy = np.abs(X[:, 1]).max()
main_ax.set_xlim(-maxx + 1, maxx + 1)
main_ax.set_ylim(-maxy + 1, maxy + 1)
main_ax.set_title("Original Data")
other_axes = [plt.subplot2grid((2, 4), (i, j)) for j in range(2, 4) for i in range(2)]
for ax, scaler in zip(
other_axes,
[StandardScaler(), RobustScaler(), MinMaxScaler(), Normalizer(norm="l2")],
):
X_ = scaler.fit_transform(X)
ax.scatter(X_[:, 0], X_[:, 1], c=y, cmap=cm2, s=60)
ax.set_xlim(-2, 2)
ax.set_ylim(-2, 2)
ax.set_title(type(scaler).__name__)
other_axes.append(main_ax)
for ax in other_axes:
ax.spines["left"].set_position("center")
ax.spines["right"].set_color("none")
ax.spines["bottom"].set_position("center")
ax.spines["top"].set_color("none")
ax.xaxis.set_ticks_position("bottom")
ax.yaxis.set_ticks_position("left")
Dataset, splitting, and baseline#
We’ll be working on Cellular Network Analysis Dataset to demonstrate these feature transformation techniques. The task is to predict network latency, given a number of features from the collected metrics
train_df, test_df = train_test_split(
pd.read_csv(
"https://raw.githubusercontent.com/WSU-AI-CyberSecurity/data/master/signal_metrics.csv"
),
test_size=0.1,
random_state=123,
)
display(train_df.head())
| Timestamp | Locality | Latitude | Longitude | Signal Strength (dBm) | Signal Quality (%) | Data Throughput (Mbps) | Latency (ms) | Network Type | BB60C Measurement (dBm) | srsRAN Measurement (dBm) | BladeRFxA9 Measurement (dBm) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11286 | 2023-05-29 23:44:54.012478 | Patliputra Colony | 25.573834 | 85.174773 | -88.823204 | 0.0 | 2.633925 | 118.470369 | LTE | -90.679320 | -97.310913 | -91.341579 |
| 11445 | 2023-05-30 08:01:00.429985 | Anandpuri | 25.459679 | 85.100593 | -84.338977 | 0.0 | 8.230689 | 62.104584 | 4G | -85.669232 | -90.443197 | -83.662386 |
| 8991 | 2023-05-25 00:24:06.665443 | Kidwaipuri | 25.718163 | 85.164846 | -95.165973 | 0.0 | 27.416884 | 39.535036 | 5G | -92.364076 | -101.027782 | -92.289394 |
| 13062 | 2023-06-02 20:06:19.279726 | Rajendra Nagar | 25.646589 | 85.123178 | -82.277196 | 0.0 | 2.681432 | 128.228075 | LTE | -78.858775 | -89.006250 | -84.283168 |
| 4151 | 2023-05-14 12:42:29.428123 | Kidwaipuri | 25.687754 | 85.008608 | -89.665566 | 0.0 | 4.607886 | 66.139469 | 4G | -90.044765 | -96.717788 | -87.254787 |
Let’s have a look at our dataset attributes and the kind of features that it contains:
train_df.info()
<class 'pandas.core.frame.DataFrame'>
Index: 15146 entries, 11286 to 15725
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Timestamp 15146 non-null object
1 Locality 15146 non-null object
2 Latitude 15146 non-null float64
3 Longitude 15146 non-null float64
4 Signal Strength (dBm) 15146 non-null float64
5 Signal Quality (%) 15146 non-null float64
6 Data Throughput (Mbps) 15146 non-null float64
7 Latency (ms) 15146 non-null float64
8 Network Type 15146 non-null object
9 BB60C Measurement (dBm) 15146 non-null float64
10 srsRAN Measurement (dBm) 15146 non-null float64
11 BladeRFxA9 Measurement (dBm) 15146 non-null float64
dtypes: float64(9), object(3)
memory usage: 1.5+ MB
train_df = train_df.drop(columns=["Timestamp", "Locality"])
test_df = test_df.drop(columns=["Timestamp", "Locality"])
EDA#
train_df.info()
<class 'pandas.core.frame.DataFrame'>
Index: 15146 entries, 11286 to 15725
Data columns (total 10 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Latitude 15146 non-null float64
1 Longitude 15146 non-null float64
2 Signal Strength (dBm) 15146 non-null float64
3 Signal Quality (%) 15146 non-null float64
4 Data Throughput (Mbps) 15146 non-null float64
5 Latency (ms) 15146 non-null float64
6 Network Type 15146 non-null object
7 BB60C Measurement (dBm) 15146 non-null float64
8 srsRAN Measurement (dBm) 15146 non-null float64
9 BladeRFxA9 Measurement (dBm) 15146 non-null float64
dtypes: float64(9), object(1)
memory usage: 1.3+ MB
We have one categorical feature and all other features are numeric features.
train_df.describe()
| Latitude | Longitude | Signal Strength (dBm) | Signal Quality (%) | Data Throughput (Mbps) | Latency (ms) | BB60C Measurement (dBm) | srsRAN Measurement (dBm) | BladeRFxA9 Measurement (dBm) | |
|---|---|---|---|---|---|---|---|---|---|
| count | 15146.000000 | 15146.000000 | 15146.000000 | 15146.0 | 15146.000000 | 15146.000000 | 15146.000000 | 15146.000000 | 15146.000000 |
| mean | 25.594919 | 85.137158 | -90.060175 | 0.0 | 16.116261 | 101.407733 | -68.567118 | -74.165153 | -68.567884 |
| std | 0.089658 | 0.090057 | 5.399430 | 0.0 | 25.620021 | 55.980000 | 40.197141 | 43.377956 | 40.147603 |
| min | 25.414575 | 84.957936 | -113.082820 | 0.0 | 1.000423 | 10.019527 | -115.667514 | -121.598760 | -114.683401 |
| 25% | 25.523497 | 85.064075 | -93.595083 | 0.0 | 2.001390 | 50.430155 | -94.022080 | -101.224898 | -93.744037 |
| 50% | 25.595584 | 85.138149 | -89.641736 | 0.0 | 2.992287 | 100.451793 | -89.112998 | -96.801157 | -89.263101 |
| 75% | 25.667371 | 85.209626 | -86.117291 | 0.0 | 9.933834 | 150.029733 | 0.000000 | 0.000000 | 0.000000 |
| max | 25.773648 | 85.316994 | -74.644848 | 0.0 | 99.985831 | 199.991081 | 0.000000 | 0.000000 | 0.000000 |
## (optional)
train_df.hist(bins=50, figsize=(20, 15))
array([[<Axes: title={'center': 'Latitude'}>,
<Axes: title={'center': 'Longitude'}>,
<Axes: title={'center': 'Signal Strength (dBm)'}>],
[<Axes: title={'center': 'Signal Quality (%)'}>,
<Axes: title={'center': 'Data Throughput (Mbps)'}>,
<Axes: title={'center': 'Latency (ms)'}>],
[<Axes: title={'center': 'BB60C Measurement (dBm)'}>,
<Axes: title={'center': 'srsRAN Measurement (dBm)'}>,
<Axes: title={'center': 'BladeRFxA9 Measurement (dBm)'}>]],
dtype=object)
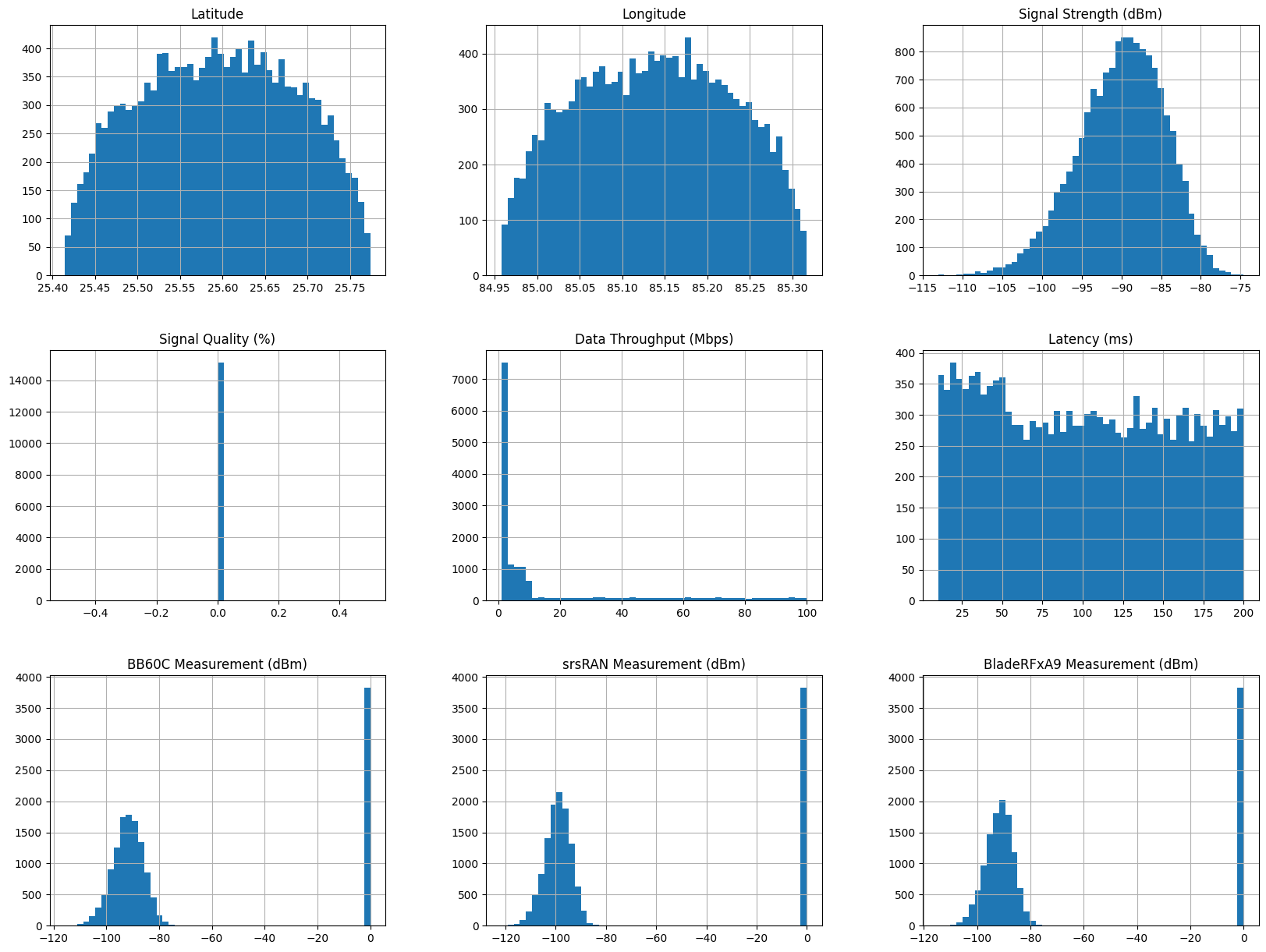
fig = pe.scatter_mapbox(
train_df[train_df["Network Type"] == "5G"],
lat="Latitude",
lon="Longitude",
mapbox_style="open-street-map",
opacity=0.2,
color="Signal Strength (dBm)",
# color='BladeRFxA9 Measurement (dBm)',
)
fig.update_layout()
fig.update_layout(margin={"r": 0, "t": 0, "l": 0, "b": 0})
fig.show("png")
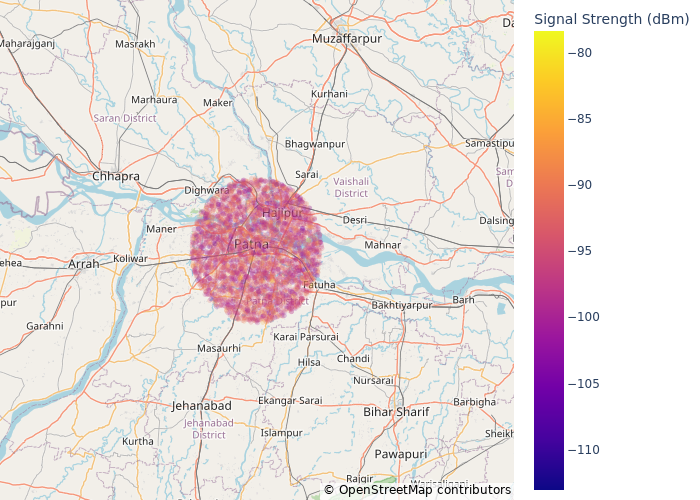
This visualisation indicates that all of the collected metrics are within certain radius of some area
fig = pe.bar(
data_frame=train_df.groupby(["Network Type"]).mean().reset_index(),
x="Network Type",
y=["Signal Strength (dBm)", "Data Throughput (Mbps)", "Latency (ms)"],
barmode="group",
title="Mean attributes grouped-by Netrowk Types (3G, 4G, 5G, LTE)",
).show("png")
/home/soraxas/micromamba/envs/wsu/lib/python3.9/site-packages/plotly/express/_core.py:2065: FutureWarning:
When grouping with a length-1 list-like, you will need to pass a length-1 tuple to get_group in a future version of pandas. Pass `(name,)` instead of `name` to silence this warning.
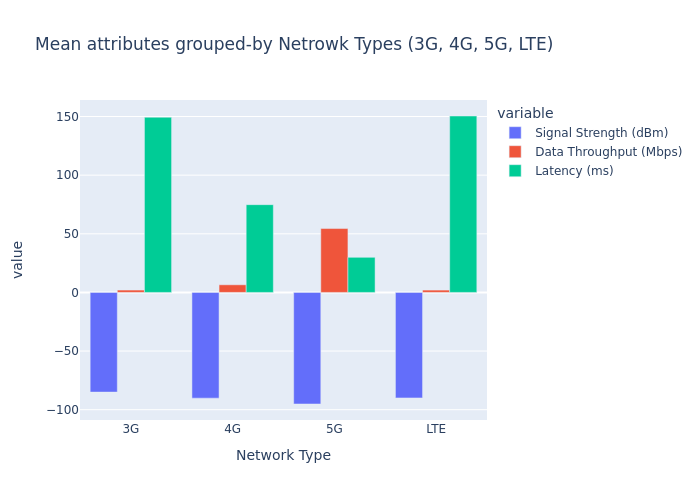
What all transformations we need to apply on the dataset?#
Here is what we see from the EDA.
Scales are quite different across columns.
Categorical variable
Network Type
Read about preprocessing techniques implemented in scikit-learn.
X_train = train_df.drop(columns=["Latency (ms)", "Network Type"])
y_train = train_df["Latency (ms)"]
X_test = test_df.drop(columns=["Latency (ms)", "Network Type"])
y_test = test_df["Latency (ms)"]
Let’s first run our baseline model DummyRegressor#
results_dict = {} # dictionary to store our results for different models
def mean_std_cross_val_scores(model, X_train, y_train, **kwargs):
"""
Returns mean and std of cross validation
Parameters
----------
model :
scikit-learn model
X_train : numpy array or pandas DataFrame
X in the training data
y_train :
y in the training data
Returns
----------
pandas Series with mean scores from cross_validation
"""
scores = cross_validate(model, X_train, y_train, **kwargs)
mean_scores = pd.DataFrame(scores).mean()
std_scores = pd.DataFrame(scores).std()
out_col = []
for i in range(len(mean_scores)):
out_col.append((f"%0.3f (+/- %0.3f)" % (mean_scores[i], std_scores[i])))
return pd.Series(data=out_col, index=mean_scores.index)
y_train
11286 118.470369
11445 62.104584
8991 39.535036
13062 128.228075
4151 66.139469
...
96 31.932823
13435 109.640342
7763 101.868511
15377 21.659666
15725 177.697604
Name: Latency (ms), Length: 15146, dtype: float64
dummy = DummyRegressor(strategy="median")
results_dict["dummy"] = mean_std_cross_val_scores(
dummy, X_train, y_train, return_train_score=True
)
/tmp/ipykernel_34641/4158382658.py:26: FutureWarning:
Series.__getitem__ treating keys as positions is deprecated. In a future version, integer keys will always be treated as labels (consistent with DataFrame behavior). To access a value by position, use `ser.iloc[pos]`
pd.DataFrame(results_dict)
| dummy | |
|---|---|
| fit_time | 0.001 (+/- 0.001) |
| score_time | 0.000 (+/- 0.000) |
| test_score | -0.001 (+/- 0.000) |
| train_score | -0.000 (+/- 0.000) |
Scaling#
This problem affects a large number of ML methods.
A number of approaches to this problem. We are going to look into the most popular ones.
Approach |
What it does |
How to update \(X\) (but see below!) |
sklearn implementation |
|---|---|---|---|
standardization |
sets sample mean to \(0\), s.d. to \(1\) |
|
scaler = StandardScaler()
X_train_scaled = scaler.fit_transform(X_train)
X_test_scaled = scaler.transform(X_test)
pd.DataFrame(X_train_scaled, columns=X_train.columns)
| Latitude | Longitude | Signal Strength (dBm) | Signal Quality (%) | Data Throughput (Mbps) | BB60C Measurement (dBm) | srsRAN Measurement (dBm) | BladeRFxA9 Measurement (dBm) | |
|---|---|---|---|---|---|---|---|---|
| 0 | -0.235178 | 0.417697 | 0.229100 | 0.0 | -0.526260 | -0.550112 | -0.533601 | -0.567268 |
| 1 | -1.508445 | -0.406029 | 1.059628 | 0.0 | -0.307800 | -0.425470 | -0.375273 | -0.375988 |
| 2 | 1.374633 | 0.307469 | -0.945649 | 0.0 | 0.441100 | -0.592026 | -0.619290 | -0.590877 |
| 3 | 0.576312 | -0.155235 | 1.441492 | 0.0 | -0.524405 | -0.256038 | -0.342146 | -0.391451 |
| 4 | 1.035463 | -1.427470 | 0.073086 | 0.0 | -0.449209 | -0.534325 | -0.519927 | -0.465470 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 15141 | -1.249304 | 0.776592 | -1.054591 | 0.0 | 2.334350 | -0.768915 | -0.626568 | -0.693106 |
| 15142 | 0.476210 | -0.607609 | -0.557700 | 0.0 | -0.557237 | -0.696676 | -0.616269 | -0.633682 |
| 15143 | -0.753742 | -1.671379 | 0.533930 | 0.0 | -0.585827 | -0.485844 | -0.451067 | -0.472370 |
| 15144 | 1.234431 | -0.034641 | -2.463528 | 0.0 | 0.052647 | -0.826386 | -0.863319 | -0.896739 |
| 15145 | 1.171396 | 0.741267 | 0.508109 | 0.0 | -0.578728 | 1.705827 | 1.709799 | 1.707951 |
15146 rows × 8 columns
knn = KNeighborsRegressor()
knn.fit(X_train_scaled, y_train)
knn.score(X_train_scaled, y_train)
0.7797007360451268
Big difference in the KNN training performance after scaling the data.
But we saw last week that training score doesn’t tell us much. We should look at the cross-validation score.
Feature transformations and the golden rule#
Let’s try cross-validation with transformed data.
knn = KNeighborsRegressor()
scaler = StandardScaler()
scaler.fit(X_train)
X_train_scaled = scaler.transform(X_train)
X_test_scaled = scaler.transform(X_test)
scores = cross_validate(knn, X_train_scaled, y_train, return_train_score=True)
pd.DataFrame(scores)
| fit_time | score_time | test_score | train_score | |
|---|---|---|---|---|
| 0 | 0.007546 | 0.027821 | 0.669859 | 0.773023 |
| 1 | 0.005107 | 0.017332 | 0.657948 | 0.774226 |
| 2 | 0.005582 | 0.019061 | 0.657192 | 0.772506 |
| 3 | 0.005142 | 0.017042 | 0.646960 | 0.775659 |
| 4 | 0.005085 | 0.017823 | 0.647245 | 0.774178 |
Do you see any problem here?
Are we applying
fit_transformon train portion andtransformon validation portion in each fold?Here you might be allowing information from the validation set to leak into the training step.
You need to apply the SAME preprocessing steps to train/validation.
With many different transformations and cross validation the code gets unwieldy very quickly.
Likely to make mistakes and “leak” information.
In these examples our test accuracies look fine, but our methodology is flawed.
Implications can be significant in practice!
Pipelines#
Can we do this in a more elegant and organized way?
YES!! Using
scikit-learn Pipeline.scikit-learn Pipelineallows you to define a “pipeline” of transformers with a final estimator.
Let’s combine the preprocessing and model with pipeline
### Simple example of a pipeline
from sklearn.pipeline import Pipeline
pipe = Pipeline(
steps=[
("imputer", SimpleImputer(strategy="median")),
("scaler", StandardScaler()),
("regressor", KNeighborsRegressor()),
]
)
Syntax: pass in a list of steps.
The last step should be a model/classifier/regressor.
All the earlier steps should be transformers.
Alternative and more compact syntax: make_pipeline#
Shorthand for
PipelineconstructorDoes not permit naming steps
Instead the names of steps are set to lowercase of their types automatically;
StandardScaler()would be named asstandardscaler
from sklearn.pipeline import make_pipeline
pipe = make_pipeline(
SimpleImputer(strategy="median"), StandardScaler(), KNeighborsRegressor()
)
pipe.fit(X_train, y_train)
Pipeline(steps=[('simpleimputer', SimpleImputer(strategy='median')),
('standardscaler', StandardScaler()),
('kneighborsregressor', KNeighborsRegressor())])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Pipeline(steps=[('simpleimputer', SimpleImputer(strategy='median')),
('standardscaler', StandardScaler()),
('kneighborsregressor', KNeighborsRegressor())])SimpleImputer(strategy='median')
StandardScaler()
KNeighborsRegressor()
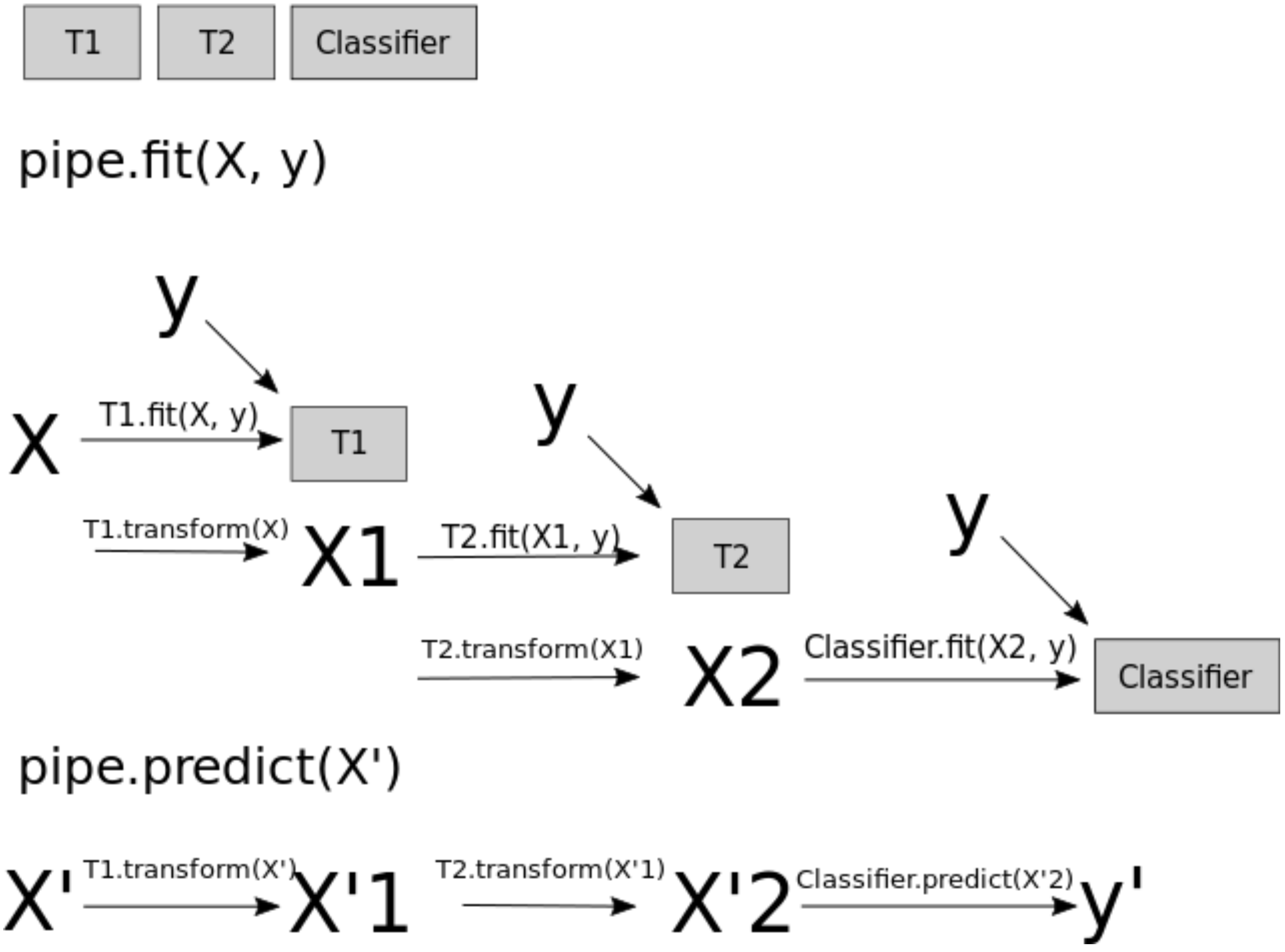
When you call fit on the pipeline, it carries out the following steps:
Fit
SimpleImputeronX_trainTransform
X_trainusing the fitSimpleImputerto createX_trainFit
StandardScaleronX_trainTransform
X_trainusing the fitStandardScalerFit the model (
KNeighborsRegressorin our case)
pipe.predict(X_train)
array([120.28679471, 75.96071876, 29.21442221, ..., 142.46425035,
44.20921845, 166.92819945])
Let’s try cross-validation with our pipeline#
results_dict["imp + scaling + knn"] = mean_std_cross_val_scores(
pipe, X_train, y_train, return_train_score=True
)
pd.DataFrame(results_dict).T
/tmp/ipykernel_34641/4158382658.py:26: FutureWarning:
Series.__getitem__ treating keys as positions is deprecated. In a future version, integer keys will always be treated as labels (consistent with DataFrame behavior). To access a value by position, use `ser.iloc[pos]`
| fit_time | score_time | test_score | train_score | |
|---|---|---|---|---|
| dummy | 0.001 (+/- 0.001) | 0.000 (+/- 0.000) | -0.001 (+/- 0.000) | -0.000 (+/- 0.000) |
| imp + scaling + knn | 0.013 (+/- 0.002) | 0.018 (+/- 0.001) | 0.656 (+/- 0.010) | 0.774 (+/- 0.001) |
Using a Pipeline takes care of applying the fit_transform on the train portion and only transform on the validation portion in each fold.
Categorical features#
Recall that we had dropped the categorical feature
Network Typefeature from the dataframe. But it could potentially be a useful feature in this task.Let’s create our
X_trainand andX_testagain by keeping the feature in the data.
X_train
| Latitude | Longitude | Signal Strength (dBm) | Signal Quality (%) | Data Throughput (Mbps) | BB60C Measurement (dBm) | srsRAN Measurement (dBm) | BladeRFxA9 Measurement (dBm) | |
|---|---|---|---|---|---|---|---|---|
| 11286 | 25.573834 | 85.174773 | -88.823204 | 0.0 | 2.633925 | -90.679320 | -97.310913 | -91.341579 |
| 11445 | 25.459679 | 85.100593 | -84.338977 | 0.0 | 8.230689 | -85.669232 | -90.443197 | -83.662386 |
| 8991 | 25.718163 | 85.164846 | -95.165973 | 0.0 | 27.416884 | -92.364076 | -101.027782 | -92.289394 |
| 13062 | 25.646589 | 85.123178 | -82.277196 | 0.0 | 2.681432 | -78.858775 | -89.006250 | -84.283168 |
| 4151 | 25.687754 | 85.008608 | -89.665566 | 0.0 | 4.607886 | -90.044765 | -96.717788 | -87.254787 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 96 | 25.482912 | 85.207093 | -95.754178 | 0.0 | 75.920386 | -99.474296 | -101.343500 | -96.393491 |
| 13435 | 25.637614 | 85.082440 | -93.071336 | 0.0 | 1.840301 | -96.570589 | -100.896749 | -94.007852 |
| 7763 | 25.527342 | 84.986643 | -87.177355 | 0.0 | 1.107868 | -88.096014 | -93.730885 | -87.531770 |
| 15377 | 25.705593 | 85.134038 | -103.361381 | 0.0 | 17.465030 | -101.784379 | -111.612911 | -104.568606 |
| 15725 | 25.699941 | 85.203912 | -87.316766 | 0.0 | 1.289739 | 0.000000 | 0.000000 | 0.000000 |
15146 rows × 8 columns
X_train = train_df.drop(columns=["Latency (ms)"])
y_train = train_df["Latency (ms)"]
X_test = test_df.drop(columns=["Latency (ms)"])
y_test = test_df["Latency (ms)"]
Let’s try to build a
KNeighborRegressoron this data using our pipeline
# pipe.fit(X_train, y_train)
This failed because we have non-numeric data.
Imagine how \(k\)-NN would calculate distances when you have non-numeric features.
Can we use this feature in the model?#
In
scikit-learn, most algorithms require numeric inputs.Decision trees could theoretically work with categorical features.
However, the sklearn implementation does not support this.
What are the options?#
Drop the column (not recommended)
If you know that the column is not relevant to the target in any way you may drop it.
We can transform categorical features to numeric ones so that we can use them in the model.
Ordinal encoding (occasionally recommended)
One-hot encoding (recommended in most cases)
One-hot encoding (OHE)#
Create new binary columns to represent our categories.
If we have \(c\) categories in our column.
We create \(c\) new binary columns to represent those categories.
Example: Imagine a language column which has the information on whether you
We can use sklearn’s
OneHotEncoderto do so.
One-hot encoding is called one-hot because only one of the newly created features is 1 for each data point.
Let’s do it on our dataset#
ohe = OneHotEncoder(sparse_output=False, dtype="int")
ohe.fit(X_train[["Network Type"]])
X_imp_ohe_train = ohe.transform(X_train[["Network Type"]])
We can look at the new features created using
categories_attribute
ohe.categories_
[array(['3G', '4G', '5G', 'LTE'], dtype=object)]
transformed_ohe = pd.DataFrame(
data=X_imp_ohe_train,
columns=ohe.get_feature_names_out(["Network Type"]),
index=X_train.index,
)
transformed_ohe
| Network Type_3G | Network Type_4G | Network Type_5G | Network Type_LTE | |
|---|---|---|---|---|
| 11286 | 0 | 0 | 0 | 1 |
| 11445 | 0 | 1 | 0 | 0 |
| 8991 | 0 | 0 | 1 | 0 |
| 13062 | 0 | 0 | 0 | 1 |
| 4151 | 0 | 1 | 0 | 0 |
| ... | ... | ... | ... | ... |
| 96 | 0 | 0 | 1 | 0 |
| 13435 | 0 | 0 | 0 | 1 |
| 7763 | 0 | 0 | 0 | 1 |
| 15377 | 0 | 0 | 1 | 0 |
| 15725 | 1 | 0 | 0 | 0 |
15146 rows × 4 columns
One-hot encoded variables are also referred to as dummy variables.
You will often see people using get_dummies method of pandas to convert categorical variables into dummy variables. That said, using sklearn’s OneHotEncoder has the advantage of making it easy to treat training and test set in a consistent way.
X_train_onehot = train_df.drop(columns=["Latency (ms)", "Network Type"])
X_train_onehot[ohe.get_feature_names_out(["Network Type"])] = ohe.transform(
train_df[["Network Type"]]
)
y_train_onehot = train_df["Latency (ms)"]
X_test_onehot = test_df.drop(columns=["Latency (ms)", "Network Type"])
X_test_onehot[ohe.get_feature_names_out(["Network Type"])] = ohe.transform(
test_df[["Network Type"]]
)
y_test_onehot = test_df["Latency (ms)"]
display(X_train_onehot.head())
display(X_test_onehot.head())
| Latitude | Longitude | Signal Strength (dBm) | Signal Quality (%) | Data Throughput (Mbps) | BB60C Measurement (dBm) | srsRAN Measurement (dBm) | BladeRFxA9 Measurement (dBm) | Network Type_3G | Network Type_4G | Network Type_5G | Network Type_LTE | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11286 | 25.573834 | 85.174773 | -88.823204 | 0.0 | 2.633925 | -90.679320 | -97.310913 | -91.341579 | 0 | 0 | 0 | 1 |
| 11445 | 25.459679 | 85.100593 | -84.338977 | 0.0 | 8.230689 | -85.669232 | -90.443197 | -83.662386 | 0 | 1 | 0 | 0 |
| 8991 | 25.718163 | 85.164846 | -95.165973 | 0.0 | 27.416884 | -92.364076 | -101.027782 | -92.289394 | 0 | 0 | 1 | 0 |
| 13062 | 25.646589 | 85.123178 | -82.277196 | 0.0 | 2.681432 | -78.858775 | -89.006250 | -84.283168 | 0 | 0 | 0 | 1 |
| 4151 | 25.687754 | 85.008608 | -89.665566 | 0.0 | 4.607886 | -90.044765 | -96.717788 | -87.254787 | 0 | 1 | 0 | 0 |
| Latitude | Longitude | Signal Strength (dBm) | Signal Quality (%) | Data Throughput (Mbps) | BB60C Measurement (dBm) | srsRAN Measurement (dBm) | BladeRFxA9 Measurement (dBm) | Network Type_3G | Network Type_4G | Network Type_5G | Network Type_LTE | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 9330 | 25.570543 | 85.083773 | -93.993151 | 0.0 | 32.993816 | -89.317574 | -102.051970 | -95.512645 | 0 | 0 | 1 | 0 |
| 9444 | 25.482447 | 85.252053 | -85.403792 | 0.0 | 1.324703 | -80.892173 | -90.616113 | -84.190972 | 0 | 0 | 0 | 1 |
| 10747 | 25.654413 | 84.995628 | -85.801469 | 0.0 | 3.386222 | -81.167757 | -91.211866 | -82.921993 | 0 | 1 | 0 | 0 |
| 14890 | 25.652035 | 84.982229 | -85.958410 | 0.0 | 2.626279 | 0.000000 | 0.000000 | 0.000000 | 1 | 0 | 0 | 0 |
| 6965 | 25.428545 | 85.090492 | -96.360206 | 0.0 | 98.067846 | -94.388360 | -105.694470 | -93.704065 | 0 | 0 | 1 | 0 |
pipe_onehot = make_pipeline(
SimpleImputer(strategy="median"), StandardScaler(), KNeighborsRegressor()
)
pipe_onehot.fit(X_train_onehot, y_train_onehot)
Pipeline(steps=[('simpleimputer', SimpleImputer(strategy='median')),
('standardscaler', StandardScaler()),
('kneighborsregressor', KNeighborsRegressor())])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Pipeline(steps=[('simpleimputer', SimpleImputer(strategy='median')),
('standardscaler', StandardScaler()),
('kneighborsregressor', KNeighborsRegressor())])SimpleImputer(strategy='median')
StandardScaler()
KNeighborsRegressor()
results_dict["imp + scaling + knn + onehot"] = mean_std_cross_val_scores(
pipe_onehot, X_train_onehot, y_train_onehot, return_train_score=True
)
pd.DataFrame(results_dict).T
/tmp/ipykernel_34641/4158382658.py:26: FutureWarning:
Series.__getitem__ treating keys as positions is deprecated. In a future version, integer keys will always be treated as labels (consistent with DataFrame behavior). To access a value by position, use `ser.iloc[pos]`
| fit_time | score_time | test_score | train_score | |
|---|---|---|---|---|
| dummy | 0.001 (+/- 0.001) | 0.000 (+/- 0.000) | -0.001 (+/- 0.000) | -0.000 (+/- 0.000) |
| imp + scaling + knn | 0.013 (+/- 0.002) | 0.018 (+/- 0.001) | 0.656 (+/- 0.010) | 0.774 (+/- 0.001) |
| imp + scaling + knn + onehot | 0.018 (+/- 0.005) | 0.023 (+/- 0.003) | 0.802 (+/- 0.004) | 0.867 (+/- 0.001) |
What did we learn with this module?#
Motivation for preprocessing
Common preprocessing steps
Imputation
Scaling
One-hot encoding
Golden rule in the context of preprocessing
Building simple supervised machine learning pipelines using
sklearn.pipeline.make_pipeline.
Problem: Different transformations on different columns#
How do we put this together with other columns in the data before fitting the regressor?
Before we fit our regressor, we want to apply different transformations on different columns
Numeric columns
imputation
scaling
Categorical columns
imputation
one-hot encoding

